Merging Data#
Summary
This section explains three ways to merge geospatial data:
Merging similar datasets using the
concat()functionMerging datasets by common attributes using the
merge()functionMerging datasets based on spatial relationships using the
sjoin()function
Merging similar datasets using concat()#
If the attributes of the input datasets are identical, they can be merged vertically using the concat() function in pandas.
Import and load required libraries:
from pathlib import Path
import geopandas as gp
import pandas as pd
Define input and output paths:
INPUT = Path.cwd().parents[0] / "00_data"
OUTPUT = Path.cwd().parents[0] / "out"
Load input datasets:
gdb_path = INPUT / "Biotopwerte_Dresden_2018.gdb"
input_data = gp.read_file(gdb_path, layer="Biotopwerte_Dresden_2018")[:1]
second_data = gp.read_file(gdb_path, layer="Biotopwerte_Dresden_2018")[1:2]
/opt/conda/envs/worker_env/lib/python3.13/site-packages/pyogrio/raw.py:198: RuntimeWarning: organizePolygons() received a polygon with more than 100 parts. The processing may be really slow. You can skip the processing by setting METHOD=SKIP, or only make it analyze counter-clock wise parts by setting METHOD=ONLY_CCW if you can assume that the outline of holes is counter-clock wise defined
return ogr_read(
The concat() function is used to concatenate two datasets. The ignore_index parameter determines whether a new index is created or if the original indexes are preserved.
ignore_index=True→ Creates a new index for the features in the merged dataset.ignore_index=False→ Preserves original indexes (which may lead to duplicate indexes).
merged_dataset = pd.concat(
[input_data, second_data],
ignore_index=True)
Check the results:
merged_dataset
| CLC_st1 | Biotpkt2018 | Shape_Length | Shape_Area | geometry | |
|---|---|---|---|---|---|
| 0 | 122 | 5.271487 | 210.523801 | 3371.947771 | MULTIPOLYGON (((415775.635 5650481.473, 415776... |
| 1 | 122 | 5.271487 | 31.935928 | 50.075513 | MULTIPOLYGON (((417850.525 5650376.33, 417846.... |
Merging based on common attributes using merge()#
If two datasets share a common attribute, they can be merged into one dataset horizontally. In this case, the output will include only rows with matching values in the common attribute.
For example, if both datasets contain an ID column, only the rows with the same ID will be merged horizontally, combining all attributes for that ID.
How the how parameter works in the merge() function
The how parameter controls how datasets are merged:
inner: The result includes only matching rows. Common attribute must be the same in both datasets from both datasets.outer: The result all rows from both datasets. Missing values are filled withNaN.left: The result includes all rows from the first dataset and matching data from the second. Non-matching rows getNaN.right: The result includes all rows from the second dataset and matching data from the first. Non-matching rows getNaN.
For more information, check the the GeoPandas documentation.
Examples how different how options affect merging with tables
The following examples show how two datasets are merged, defined by the how parameter.
Dataset 1:
ID |
Type |
|---|---|
1 |
A |
3 |
C |
6 |
D |
Dataset 2:
ID |
Time |
|---|---|
1 |
15 min |
3 |
16 min |
4 |
17 min |
inner: Keeps only the rows with matchingIDvalues in both datasets.
ID |
Type |
Time |
|---|---|---|
1 |
A |
15 min |
3 |
C |
16 min |
outer: Includes all rows from both datasets; missing values are filled withNaN.
ID |
Type |
Time |
|---|---|---|
1 |
A |
15 min |
3 |
C |
16 min |
6 |
D |
NaN |
4 |
NaN |
17 min |
left: Keeps all rows from Dataset 1 and adds matching values from Dataset 2; unmatched values getNaN.
ID |
Type |
Time |
|---|---|---|
1 |
A |
15 min |
3 |
C |
16 min |
6 |
D |
NaN |
right: Keeps all rows from Dataset 2 and adds matching values from Dataset 1; unmatched values getNaN.
ID |
Type |
Time |
|---|---|---|
1 |
A |
15 min |
3 |
C |
16 min |
4 |
NaN |
17 min |
Code example: Using the merge() function in Python
If the common attribute (column) has the same name in both datasets:
Merge based on the column CLC_st1 which is common in both datasets:
on: Defines the similarity attribute.how: Controls how datasets are merged.
merged_dataset = input_data.merge(
second_data,
on='CLC_st1',
how='inner')
Then check the result with the print function:
merged_dataset.T
| 0 | |
|---|---|
| CLC_st1 | 122 |
| Biotpkt2018_x | 5.271487 |
| Shape_Length_x | 210.523801 |
| Shape_Area_x | 3371.947771 |
| geometry_x | MULTIPOLYGON (((415775.6353000002 5650481.4728... |
| Biotpkt2018_y | 5.271487 |
| Shape_Length_y | 31.935928 |
| Shape_Area_y | 50.075513 |
| geometry_y | MULTIPOLYGON (((417850.5251000002 5650376.3303... |
Handling duplicate column names
If both datasets have columns with the same name (other than the common attribute), GeoPandas adds suffixes _x and _y by default.
Example: If both datasets contain a column named area, the merged dataset will have area_x (from the first dataset) and area_y (from the second dataset).
Customizing suffixes#
To rename these suffixes within the merge() function, use the suffixes parameter:
In the example, FIRST added to the first dataset and SECOND added to the second dataset.
merged_dataset = input_data.merge(
second_data,
on='CLC_st1',
how='inner',
suffixes=('_FIRST', '_SECOND')) # rename suffixes
Check the result with the print function:
merged_dataset.T
| 0 | |
|---|---|
| CLC_st1 | 122 |
| Biotpkt2018_FIRST | 5.271487 |
| Shape_Length_FIRST | 210.523801 |
| Shape_Area_FIRST | 3371.947771 |
| geometry_FIRST | MULTIPOLYGON (((415775.6353000002 5650481.4728... |
| Biotpkt2018_SECOND | 5.271487 |
| Shape_Length_SECOND | 31.935928 |
| Shape_Area_SECOND | 50.075513 |
| geometry_SECOND | MULTIPOLYGON (((417850.5251000002 5650376.3303... |
Merging only selected attributes
By default, merging includes all attributes from both datasets. However, you can specify only the required attribute.
To select the specific attributes (columns) for any process, these attributes are defined in double brackets [[ ]]. The “common” attribute (on parameter), must be defined in both datasets.
Note:
Use double brackets
[[ ]]to preserve the tabular format (GeoDataFrame).Using single brackets
[ ]creates a series of data, which is not in tabular format (GeoDataFrame) and makes an error with operating themerge()function.
The following output includes only the CLC_st1 column, which is the common column in both datasets and geometry column of the second dataset.
merged_dataset = input_data[['CLC_st1']].merge(
second_data[['CLC_st1','geometry']],
on='CLC_st1',
how='inner')
merged_dataset # Check the result
| CLC_st1 | geometry | |
|---|---|---|
| 0 | 122 | MULTIPOLYGON (((417850.525 5650376.33, 417846.... |
If the common attribute (column) has different names in both datasets:
Load the datasets:
input_data = gp.read_file(INPUT / "Biotopwerte_Dresden_2018.gdb")
second_data = gp.read_file(INPUT / "clc_legend.csv")
Check the columns:
The output shows that both datasets include the CLC code but with different names:
CLC_st1in the first datasetCLC_CODEin the second dataset
input_data.columns
Index(['CLC_st1', 'Biotpkt2018', 'Shape_Length', 'Shape_Area', 'geometry'], dtype='object')
second_data.columns
Index(['GRID_CODE', 'CLC_CODE', 'LABEL1', 'LABEL2', 'LABEL3', 'RGB'], dtype='object')
Merge using the merge() function:
In the example below, the CLC codes in the input dataset are labaled using the LABEL3 column in the second dataset.
merged_dataset = input_data.merge(
second_data[["CLC_CODE", "LABEL3"]],
left_on ='CLC_st1',
right_on ='CLC_CODE',
how ='inner')
merged_dataset # Check the result
| CLC_st1 | Biotpkt2018 | Shape_Length | Shape_Area | geometry | CLC_CODE | LABEL3 | |
|---|---|---|---|---|---|---|---|
| 0 | 122 | 5.271487 | 210.523801 | 3371.947771 | MULTIPOLYGON (((415775.635 5650481.473, 415776... | 122 | Road and rail networks and associated land |
| 1 | 122 | 5.271487 | 31.935928 | 50.075513 | MULTIPOLYGON (((417850.525 5650376.33, 417846.... | 122 | Road and rail networks and associated land |
| 2 | 122 | 5.271487 | 810.640513 | 1543.310127 | MULTIPOLYGON (((417886.917 5650544.364, 417909... | 122 | Road and rail networks and associated land |
| 3 | 122 | 5.271487 | 24.509066 | 36.443441 | MULTIPOLYGON (((423453.146 5650332.06, 423453.... | 122 | Road and rail networks and associated land |
| 4 | 122 | 5.271487 | 29.937138 | 40.494155 | MULTIPOLYGON (((417331.434 5650889.039, 417330... | 122 | Road and rail networks and associated land |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 33918 | 124 | 8.000000 | 9.072443 | 4.947409 | MULTIPOLYGON (((414814.645 5666810.533, 414814... | 124 | Airports |
| 33919 | 124 | 8.000000 | 1369.670301 | 63201.087919 | MULTIPOLYGON (((414791.962 5666543.765, 414803... | 124 | Airports |
| 33920 | 124 | 8.000000 | 395.094767 | 708.068118 | MULTIPOLYGON (((415006.509 5666816.796, 415004... | 124 | Airports |
| 33921 | 231 | 10.981298 | 110.373766 | 99.282910 | MULTIPOLYGON (((417478.532 5665012.465, 417477... | 231 | Pastures |
| 33922 | 231 | 10.981298 | 1401.832280 | 38939.551849 | MULTIPOLYGON (((417482.897 5665014.048, 417475... | 231 | Pastures |
33923 rows × 7 columns
Merging datasets based on spatial relationships using sjoin()#
Another way to merge datasets is by using spatial relationships instead of common attributes.
The sjoin() function can be used to join two datasets based on their spatial relationships.
Key parameters:
predicate- Defines the type of spatial relationship.how- Specifies how the datasets are combined.
Available operations with the predicate parameter
The following list explains available operations:
contains: Object A completely encloses object B (no boundary touch).covers: Object A completely contains object B (boundaries may touch).within: Object A is completely inside object B (no boundary touch).covered_by: Object A is completely within object B (boundaries may touch).touches: Objects A and B meet only at boundaries.overlaps: Objects A and B share an area.crosses: Objects A and B intersect at discrete points.intersects: Object A and B touch, cross or share an area.
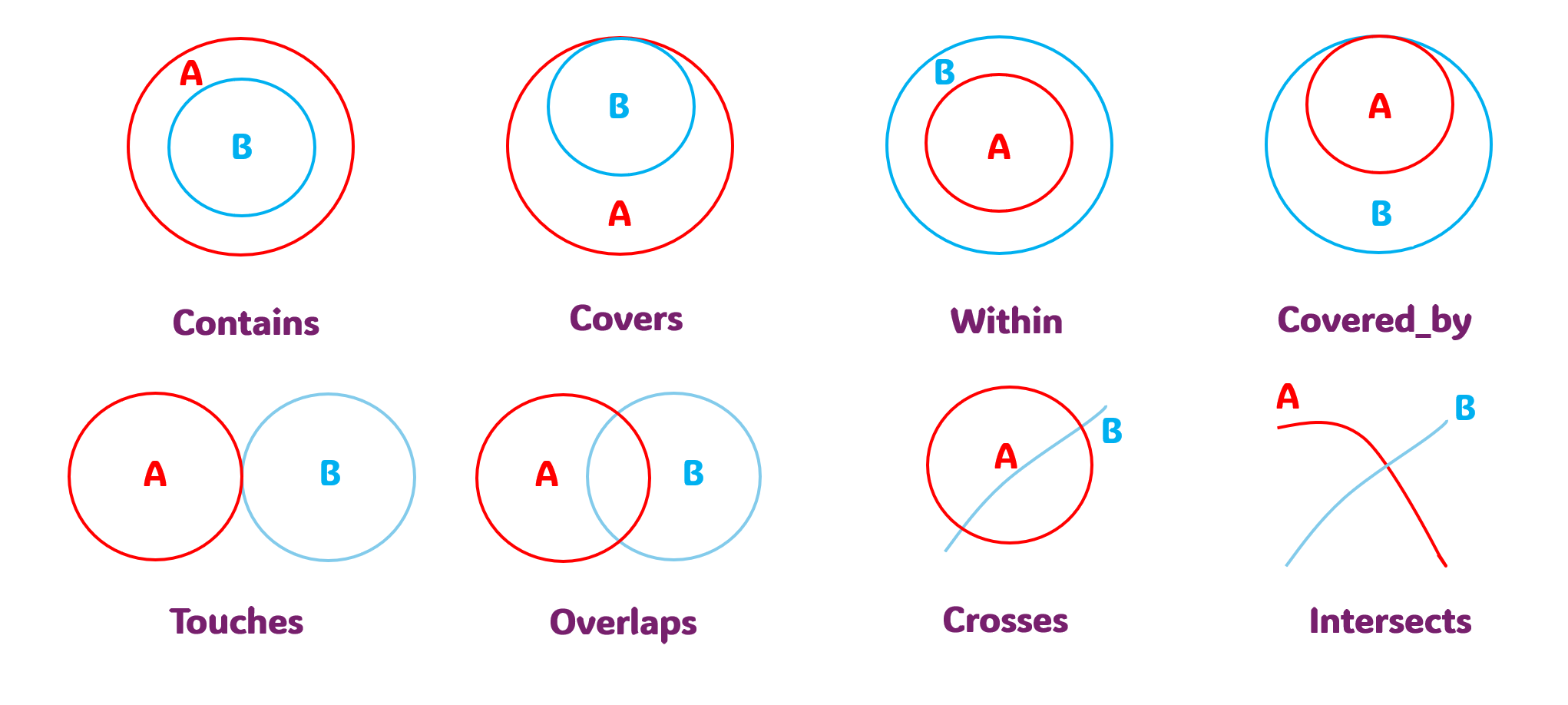
Fig. 15 Available operations with the predicate parameter#
Check the ArcGIS documentation to learn more about spatial relationships.
Available operations with the how parameter
The how parameter determines how the two datasets are merged:
left: keeps all records from the left dataset (first dataset) and adds matching records from the right dataset. The index comes from the left dataset.right: Keeps all records from the right dataset (second dataset) and adds matching records from the left dataset. The index comes from the right dataset.inner: Keeps only matching records from both datasets (where spatial conditions are met). The index comes from the right dataset.
Check GeoPandas documentation to learn more about sjoin() function.
Handling duplicate column names
If both datasets have columns with the same name, GeoPandas automatically assigns suffixes (_left and _right) to distinguish them.
To customize these suffixes, use:
lsuffix: Defines a suffix for the left dataset.rsuffix: Defines a suffix for the right dataset.
Code example: Using the sjoin() function in Python
Import and load required libraries:
from pathlib import Path
import geopandas as gp
import pandas as pd
Load the input and join datasets:
input_data = gp.read_file(gdb_path, layer="Biotopwerte_Dresden_2018")
# District data from the Dresden portal Already mentioned in the Clipping Data chapter
import requests
geojson_url = "https://kommisdd.dresden.de/net4/public/ogcapi/collections/L137/items"
response = requests.get(geojson_url)
if response.status_code == 200:
join_data = gp.read_file(geojson_url)
join_data= join_data[join_data['id'] == '99']
else:
print("Error:", response.text)
Set the same coordinate system:
join_data =join_data.to_crs(input_data.crs)
Perform a spatial join with sjoin() function and default suffixes:
The output includes the land cover and biodiversity values for the id 99 which is related to the the Altfranken/Gompitz area in Dresden.
join_result = input_data.sjoin(
join_data,
predicate='intersects',
how='inner')
Check the columns in the output:
list(join_result.columns)
['CLC_st1',
'Biotpkt2018',
'Shape_Length',
'Shape_Area',
'geometry',
'index_right',
'id',
'bez',
'bez_lang',
'flaeche_km2',
'sst',
'sst_klar',
'historie',
'aend']
Display the first 3 rows:
join_result.head(3)
| CLC_st1 | Biotpkt2018 | Shape_Length | Shape_Area | geometry | index_right | id | bez | bez_lang | flaeche_km2 | sst | sst_klar | historie | aend | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 204 | 122 | 5.271487 | 487783.286284 | 2.869516e+06 | MULTIPOLYGON (((401752.251 5661561.055, 401808... | 40 | 99 | Altfranken/Gompitz | Altfranken/Gompitz mit Ockerwitz, Pennrich, Ro... | 13.0048 | None | None | akt | 20.03.2025 00:00:00 |
| 261 | 122 | 5.271487 | 1765.048621 | 6.984237e+03 | MULTIPOLYGON (((406601.829 5652484.777, 406604... | 40 | 99 | Altfranken/Gompitz | Altfranken/Gompitz mit Ockerwitz, Pennrich, Ro... | 13.0048 | None | None | akt | 20.03.2025 00:00:00 |
| 270 | 122 | 5.271487 | 720.099258 | 2.558749e+03 | MULTIPOLYGON (((402096.709 5654422.947, 402077... | 40 | 99 | Altfranken/Gompitz | Altfranken/Gompitz mit Ockerwitz, Pennrich, Ro... | 13.0048 | None | None | akt | 20.03.2025 00:00:00 |
Perform a spatial join with sjoin() function and customizing suffixes:
join_result = input_data.sjoin(
join_data,
predicate='intersects',
how='inner',
rsuffix='_border') # Defines a suffix for the right dataset
list(join_result.columns) # Checks the columns in the output
['CLC_st1',
'Biotpkt2018',
'Shape_Length',
'Shape_Area',
'geometry',
'index__border',
'id',
'bez',
'bez_lang',
'flaeche_km2',
'sst',
'sst_klar',
'historie',
'aend']
Selecting specific columns
Instead of including all attributes from both datasets, you can specify which columns should be included in the spatial join by using double brackets [[ ]].
Since the spatial join involves geometry attributes, the geometry attribute must be defined among selected columns in both datasets.
join_result = input_data[['CLC_st1','geometry']] \
.sjoin(
join_data[['id','geometry']],
predicate='intersects', how='right')
list(join_result.columns)
['index_left', 'CLC_st1', 'id', 'geometry']
Using the gp.sjoin() function
Another way of spatial joining is to use the gp.sjoin() function from GeoPandas. The key parameters are exactly the same as in the sjoin() function, the only difference is that both datasets are passed as arguments inside the function, which is how functions generally work in Python.
joined_result= gp.sjoin(
input_data, # First dataset / left dataset
join_data, # Second dataset / right dataset
how='right',
predicate='intersects',
lsuffix='_left')
list(joined_result.columns)
['index__left',
'CLC_st1',
'Biotpkt2018',
'Shape_Length',
'Shape_Area',
'id',
'bez',
'bez_lang',
'flaeche_km2',
'sst',
'sst_klar',
'historie',
'aend',
'geometry']
Selecting specific columns
joined_result= gp.sjoin(
input_data[['CLC_st1','geometry']],
join_data[['id','geometry']],
how='right',
predicate='intersects',
lsuffix='_left')
list(joined_result.columns)
['index__left', 'CLC_st1', 'id', 'geometry']
