Clipping Data#
Clipping is a process used to extract specific parts of data based on a predefined dataset that represents the area of interest. The geopandas and matplotlib packages are used for this process.
import geopandas as gp
import matplotlib.pyplot as plt
from pathlib import Path
INPUT = Path.cwd().parents[0] / "00_data"
OUTPUT = Path.cwd().parents[0] / "out"
OUTPUT.mkdir(exist_ok=True)
gdb_path = INPUT / "Biotopwerte_Dresden_2018.gdb"
Clipping Based on Zone (Dataset)#
The areas of interest in this chapter are districts in Dresden.
Dresden Portal!
The districts are extracted from the Portal of Dresden.
For retrieving the data, from the Geojson section of the portal, based on the interested format, the URL is chosen.
geojson_url = "https://kommisdd.dresden.de/net4/public/ogcapi/collections/L137/items"
Then the requests library is imported and a request is sent to the portal to extract the data.
import requests
response = requests.get(geojson_url)
Then if the request is successful (code 200), it will load and plot the data.
if response.status_code == 200:
gdf = gp.read_file(geojson_url)
print(gdf.head())
ax=gdf.plot()
ax.set_axis_off()
else:
print("Error:", response.text)
Show code cell output
id bez \
0 02 Pirnaische Vorstadt
1 53 Striesen-Süd
2 97 Gorbitz-Nord/Neu-Omsewitz
3 22 Mickten
4 95 Gorbitz-Süd
bez_lang flaeche_km2 sst sst_klar \
0 Pirnaische Vorstadt 0.9281 None None
1 Striesen-Süd mit Johannstadt-Südost 1.3276 None None
2 Gorbitz-Nord/Neu-Omsewitz 0.8557 None None
3 Mickten mit Trachau-Süd, Übigau und Kaditz-Süd 4.2435 None None
4 Gorbitz-Süd 1.2604 None None
historie aend \
0 akt 20.03.2025 00:00:00
1 akt 20.03.2025 00:00:00
2 akt 20.03.2025 00:00:00
3 akt 20.03.2025 00:00:00
4 akt 20.03.2025 00:00:00
geometry
0 POLYGON ((13.75017 51.04619, 13.75036 51.04612...
1 POLYGON ((13.77558 51.04538, 13.77544 51.04543...
2 POLYGON ((13.6723 51.05024, 13.67231 51.0503, ...
3 POLYGON ((13.70028 51.08324, 13.70015 51.08322...
4 POLYGON ((13.66572 51.04003, 13.66609 51.04002...
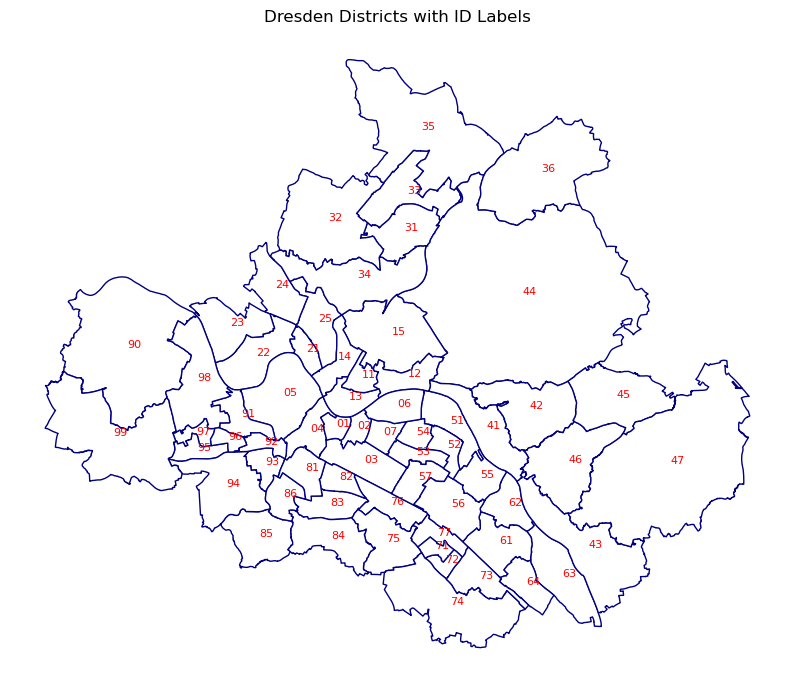
Selecting the area of interest!
To select the area of interest, the methods mentioned in the Selecting and Filtering Chapter could be used, but also it is possible to add the labels to maps and select based on labels.
To add the labels, first, the position of the labels should be defined.
The centroid of the polygons would be the position of the labels and is generated using the geometry.centroid method and stored in a new column.
gdf["centroid"] = gdf.geometry.centroid
/tmp/ipykernel_2419/1565410799.py:1: UserWarning: Geometry is in a geographic CRS. Results from 'centroid' are likely incorrect. Use 'GeoSeries.to_crs()' to re-project geometries to a projected CRS before this operation.
gdf["centroid"] = gdf.geometry.centroid
For adding the labels the text function is used.
The coordinates of the centroid are the position of the labels and the label text is defined with str.
In the following example, the IDs of the districts are used.
Labels in the map!
To customize the labels more, check here.
fig, ax = plt.subplots(figsize=(10, 10))
gdf.plot(ax=ax, color='white', edgecolor='navy')
ax.set_axis_off()
for idx, row in gdf.iterrows():
plt.text(row.centroid.x, row.centroid.y, str(row['id']),
fontsize=8,
color='red')
plt.title("Dresden Districts with ID Labels")
plt.show()
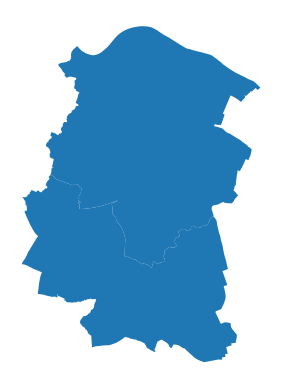
Now using the labels the interested area is selected.
clipping_dataset= gdf[gdf['id'].isin(['90','99'])]
ax=clipping_dataset.plot()
ax.set_axis_off()
The path to the original dataset and the clipping dataset, based on which the original dataset should be clipped, will be defined.
original_dataset = gp.read_file(gdb_path, layer="Biotopwerte_Dresden_2018")
/opt/conda/envs/worker_env/lib/python3.13/site-packages/pyogrio/raw.py:198: RuntimeWarning: organizePolygons() received a polygon with more than 100 parts. The processing may be really slow. You can skip the processing by setting METHOD=SKIP, or only make it analyze counter-clock wise parts by setting METHOD=ONLY_CCW if you can assume that the outline of holes is counter-clock wise defined
return ogr_read(
The coordinate system of the datasets is checked. If the coordinate systems do not match, the clipping process will not proceed.
original_dataset.crs
<Projected CRS: EPSG:25833>
Name: ETRS89 / UTM zone 33N
Axis Info [cartesian]:
- E[east]: Easting (metre)
- N[north]: Northing (metre)
Area of Use:
- name: Europe between 12°E and 18°E: Austria; Croatia; Denmark - offshore and offshore; Germany - onshore and offshore; Italy - onshore and offshore; Norway including Svalbard - onshore and offshore.
- bounds: (12.0, 34.79, 18.01, 84.01)
Coordinate Operation:
- name: UTM zone 33N
- method: Transverse Mercator
Datum: European Terrestrial Reference System 1989 ensemble
- Ellipsoid: GRS 1980
- Prime Meridian: Greenwich
clipping_dataset.crs
<Geographic 2D CRS: EPSG:4326>
Name: WGS 84
Axis Info [ellipsoidal]:
- Lat[north]: Geodetic latitude (degree)
- Lon[east]: Geodetic longitude (degree)
Area of Use:
- name: World.
- bounds: (-180.0, -90.0, 180.0, 90.0)
Datum: World Geodetic System 1984 ensemble
- Ellipsoid: WGS 84
- Prime Meridian: Greenwich
If the coordinate systems are not the same, they are transformed to match.
clipping_dataset = clipping_dataset.to_crs('EPSG:25833')
The dataset is then clipped by defining the original dataset and the clipping dataset using the clip function in the geopandas package.
clipped_dataset = gp.clip(original_dataset, clipping_dataset)
And the result of the clip operation will be printed out.
clipped_dataset.head(3)
| CLC_st1 | Biotpkt2018 | Shape_Length | Shape_Area | geometry | |
|---|---|---|---|---|---|
| 261 | 122 | 5.271487 | 1765.048621 | 6984.236836 | POLYGON ((404092.96 5654379.53, 404098.204 565... |
| 5928 | 322 | 18.055116 | 204.848619 | 582.329680 | POLYGON ((404867.05 5654106.356, 404862.483 56... |
| 13027 | 142 | 7.000000 | 481.434416 | 11886.564349 | POLYGON ((405339.13 5654136.481, 405172.058 56... |
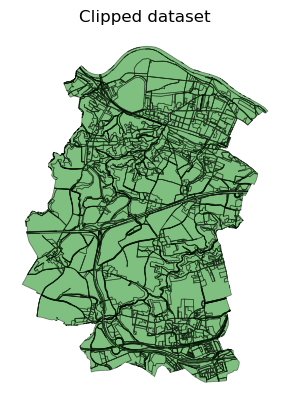
ax = clipped_dataset.plot(color="green", linewidth=0.5, alpha=0.5, edgecolor="black")
ax.set_title("Clipped dataset")
ax.set_axis_off()
The result can be saved in various formats. All you need to do is define the output path and export based on the format you are interested in.
Shapefile Format
output_shp = OUTPUT / "clipped.shp"
clipped_dataset.to_file(output_shp)
/tmp/ipykernel_2419/1343647297.py:2: UserWarning: Column names longer than 10 characters will be truncated when saved to ESRI Shapefile.
clipped_dataset.to_file(output_shp)
/opt/conda/envs/worker_env/lib/python3.13/site-packages/pyogrio/raw.py:723: RuntimeWarning: Normalized/laundered field name: 'Biotpkt2018' to 'Biotpkt201'
ogr_write(
/opt/conda/envs/worker_env/lib/python3.13/site-packages/pyogrio/raw.py:723: RuntimeWarning: Normalized/laundered field name: 'Shape_Length' to 'Shape_Leng'
ogr_write(
GeoPackage Format
output_gdb = OUTPUT / "clipped.gpkg"
clipped_dataset.to_file(
output_gdb,
layer="clipped_dataset",
driver="GPKG")
CSV Format or WKT Format
Geometry as WKT
Our data is already in WKT format. If the dataset doesn’t have a geometry column in Well Known Text format, it should be converted first, as mentioned in the next steps. You will see in the final CSV file saved below that the two columns “geometry” and “geometry_wkt” match.
Have a look at the geometry column, which commonly exists for GeoDataFrames
clipped_dataset.geometry
Show code cell output
261 POLYGON ((404092.96 5654379.53, 404098.204 565...
5928 POLYGON ((404867.05 5654106.356, 404862.483 56...
13027 POLYGON ((405339.13 5654136.481, 405172.058 56...
15142 POLYGON ((405337.399 5654208.987, 405336.192 5...
3572 POLYGON ((405336.192 5654214.027, 405335.975 5...
...
24741 POLYGON ((403517.379 5661823.326, 403517.089 5...
11147 POLYGON ((403506.01 5661746.878, 403512.79 566...
33830 MULTIPOLYGON (((403834.523 5661937.043, 403787...
33829 POLYGON ((403103.13 5662019.63, 403184.07 5662...
33831 POLYGON ((403506.092 5662087.066, 403484.425 5...
Name: geometry, Length: 2923, dtype: geometry
First, ensure that the geometry column contains valid geometries. We need to check if all values in clipped_dataset["geometry"] are actual shapely.geometry objects.
Check if there are any missing geometries (.isna()).
print(clipped_dataset["geometry"].isna().sum()) # Count missing values
0
Check the types of geometries.
print(clipped_dataset.geom_type.value_counts())
Polygon 2833
MultiPolygon 90
Name: count, dtype: int64
Explicitly store WKT geometries in a new column. This uses the apply function with a lambda expression to convert each geometry in the column to a string format.
from shapely.geometry import Polygon, MultiPolygon
clipped_dataset["geometry_wkt"] = clipped_dataset["geometry"].apply(
lambda geom: geom.wkt if isinstance(geom, (Polygon, MultiPolygon)) else None
)
The output path is defined, and the data is saved as a CSV file. If the index column, which represents unique values for rows, is not needed, the index parameter can be set to False.
Parameters for CSV file
To learn how to customize your CSV data, visit the pandas DataFrame to CSV page.
csv_path = OUTPUT / "clipped_dataset.csv"
clipped_dataset.to_csv(csv_path, index=True)
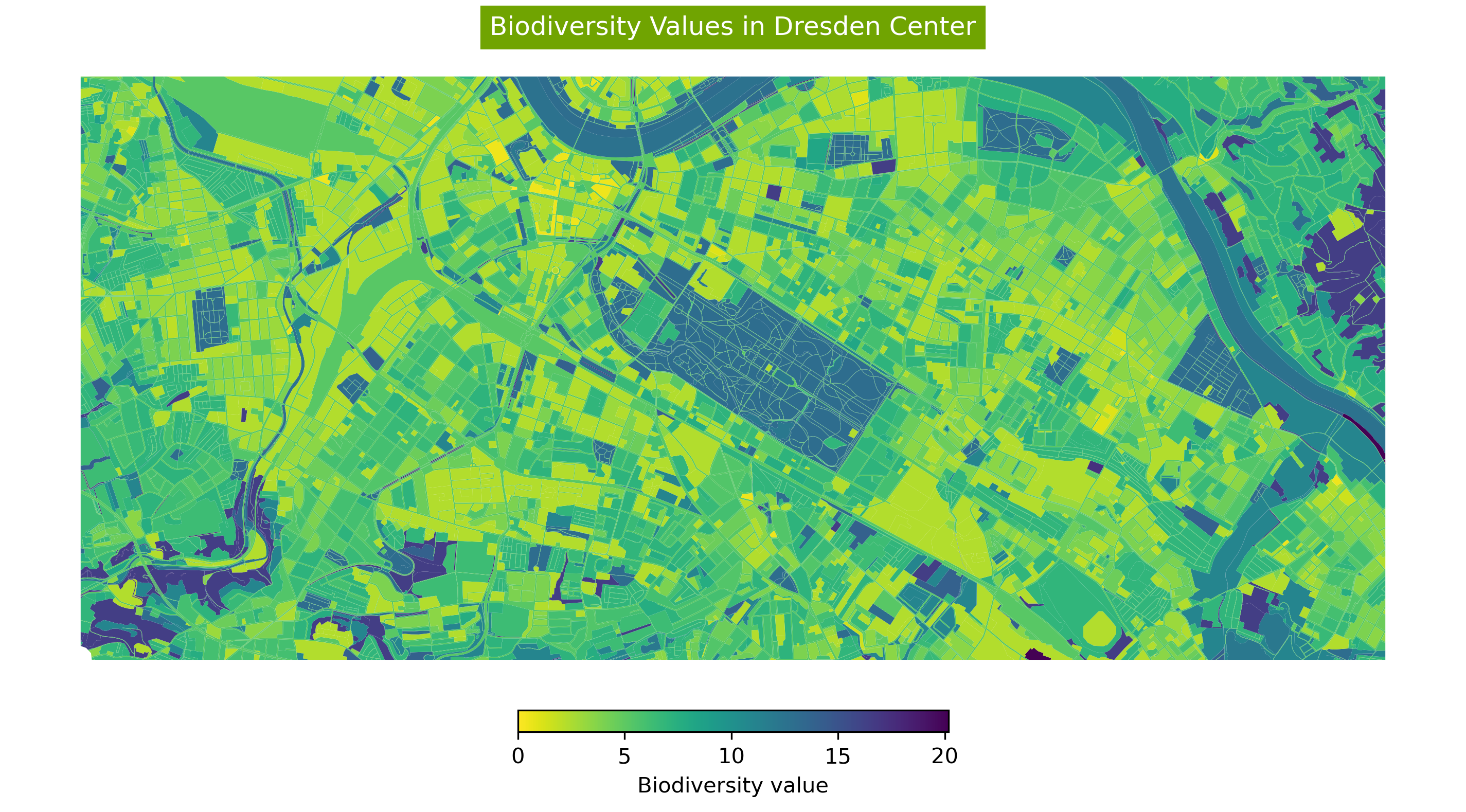
Clipping Based on Bounding Box#
To show a specific area of the dataset, the data can be clipped to a defined bounding box and then visualized.
For this reason, the required part of the dataset is selected and the bounding box is extracted for this rows.
Bounding Box
The bounding box extraction explained more in Creating Maps Chapter.
required_part= original_dataset[80:120].total_bounds
Then the dataset clipped for plotting:
gdf_clipped = gp.clip(original_dataset, required_part)
fig, ax = plt.subplots(
nrows=1, ncols=1, figsize=(12, 18), dpi=300)
ax = gdf_clipped.plot(
ax=ax,
column='Biotpkt2018',
cmap='viridis_r',
legend=True,
legend_kwds={
"label": "Biodiversity value", "orientation": "horizontal",
"shrink": 0.3, "pad": 0.01})
ax.set_title('Biodiversity Values in Dresden Center',
backgroundcolor='#70A401',
color='white')
ax.set_axis_off()